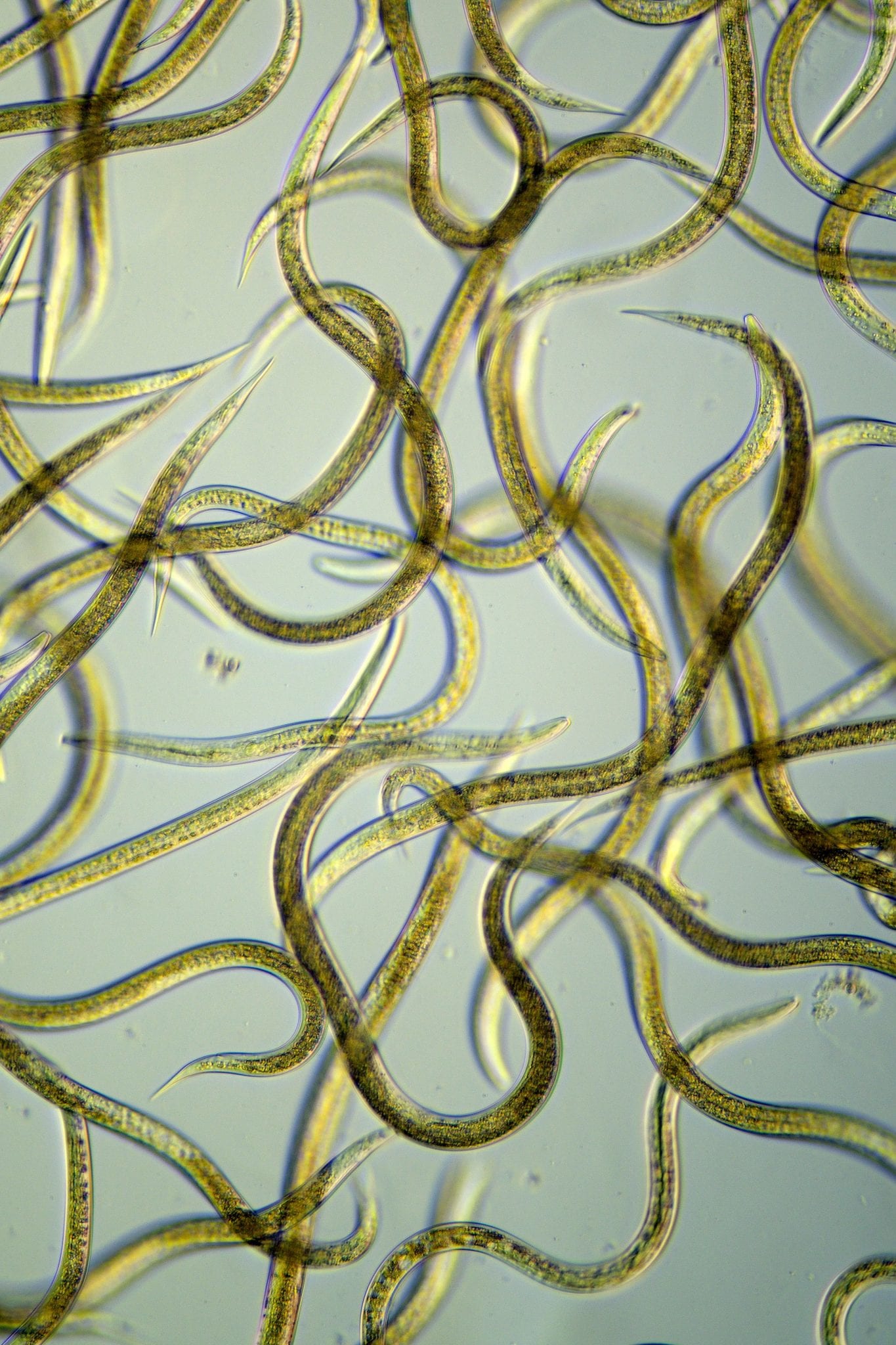
Nematodes or roundworms are microscopic, non-segmented, worm-like organisms found in all sorts of ecosystems, from deep ocean vents to mountain peaks. There is even a beer-mat nematode. They may be predatory, parasitic (animals and humans), herbivorous or feed on fungi or bacteria.
What do nematodes do?
Plant-parasitic nematodes are extremely damaging to food crops in all regions of the world and have been estimated to cause a 14% loss of crop yields globally. On the other hand, nematode species that infect insects and slugs (entomopathogenic nematodes) have long been used to provide an environmentally safe form of pest control. Many nematodes also play an important role in the nutrient cycles of soils, through excretion and the decomposition of their bodies. Additionally, they can influence carbon fluxes, the structure of soil food webs and even the diversity of plants in a given location.
How do nematodes relate to soil health?
Nematodes are abundant and ubiquitous in all soils, including those used for crop production. In fact, one square metre of field soil could contain tens of millions of nematodes of many different types. Some nematodes will reproduce just once, with many offspring, while others, especially if in stable environments, will tend to live longer and produce fewer offspring but will reproduce more than once. The abundance and ratio of these different types of nematode can provide a useful tool for measuring the condition of the soil ecosystem, and many indices that explore species richness, diversity and maturity have been developed for this purpose.
To do this, the type and quantity of nematodes needs to be determined. This is far from simple, requires a high level of taxonomic expertise and can be very time-consuming. For instance, a 100 g soil sample may contain hundreds of individuals and take more than two hours to examine.
DNA barcoding
The alternative is to use identification methods that require less expert knowledge. One such method is DNA barcoding. This examines key areas of an organism’s DNA for identification and can be done by anyone trained in PCR and DNA sequencing.
DNA barcoding of nematodes, like any organism, starts with building a reference database populated with the DNA barcode for as many species as possible. This first step is crucial: a high level of accuracy is needed to ensure success. In other words, any DNA barcode entered into such a database must be from a carefully and accurately visually identified individual. From that point, identifying an unknown individual is simply a matter of sequencing its DNA barcode region and comparing it to the available barcodes in the reference database.
Where are we now with this technology?
When DNA barcoding was introduced in 2003, the available sequencing technology only allowed for the barcoding of one individual at a time. Since then the development of new high-throughput sequencing technologies has opened up a whole world of possibilities with their capacity to sequence multiple individuals and samples in a single run, known as metabarcoding.
This technology means that all nematodes extracted from a sample can be bulked together with those from several other samples and sequenced in one go. DNA metabarcoding typically involves extracting nematodes from each sample, bulking them together in a drop of water, performing DNA extraction, and then isolating and making copies of the barcode region from the bulk DNA. This is then sequenced, and through bioinformatics, the source of each barcode is identified by matching it against all barcodes in a reference database. The resulting DNA barcodes allows each constituent species to be detected. Furthermore, the number of DNA barcodes recovered for each species in a sample should, theoretically, indicate its relative abundance in the sample.
Despite this, the use of metabarcoding in soil quality assessment is still limited. The prediction of nematode quantities in bulk samples from the number of DNA barcodes is not yet accurate enough to be useful. To fully substitute the traditional method, this alternative approach needs to be capable of not only detecting the groups or types present in a sample, but also correctly predicting the abundances of these groups.
What is the future for this technology?
Research is still ongoing with the goal to better understand how to better predict abundances of nematode groups from the abundances of their DNA barcodes. If it succeeds, work that currently take days could be done in a matter of hours with DNA metabarcoding. More importantly, it will be much more accessible as lack of taxonomic expertise will no longer prevent the use of nematodes as bioindicators in small or even large-scale soil health assessment studies.
Dr Mohammed Ahmed, a specialist in nematode taxonomy and phylogenetics, examined DNA barcoding of nematode communities for his PhD from Harper Adams University. He is currently based at the Swedish Museum of Natural History (Stockholm). Dr Matthew Back is a Reader in Nematology at Harper Adams University with interests in plant nematology, disease complexes involving nematodes, and the management of soil-borne diseases. Follow Matthew Back on Twitter
If you have any questions about becoming a member of CHAP, or would like to reach out to the CHAP team to discuss other project opportunities, please email us at enquiries@chap-solutions.co.uk
Please note, the opinions expressed in this article are the author’s own and do not necessarily reflect the views or opinions of CHAP.